
Wormhole: Ortholog Prediction
through Machine Learning

|
Wormhole: Ortholog Prediction |
This page provides information about how to use the WORMHOLE ortholog prediction web tool and how to interpret the information provided. The following sections describe the different components of the search tool and what options are available for each component.
WORMHOLE uses multilayer machine learning to predict novel least diverged orthologs (LDOs) by integrating predictions from 17 ortholog prediction algorithms. WORMHOLE employs support vectors machine (SVM) classifiers, a type of supervised machine learning model, trained on a reference set of well-defined LDOs (the PANTHER LDOs). These models classify each novel gene pair as either an LDO or non-LDO based on the pattern of predictions made by the 17 input algorithms. The weight given to each algorithm is learned based on the performance of that algorithm and identifying PANTHER LDOs.
Once trained, the WORMOLE SVM classifiers were applied to every predicted ortholog pair between six examined genomes (yeast, nematodes, fruit flies, zebrafish, humans, and mice) in order to generate confidence scores. These scores range from 0 to 1 and reflect the confidence that the predicted gene pair is an LDO, with high scores indicating high confidence and low score low confidence. The scores are scaled such that a score of 0.5 returns maximum performance across the PANTHER LDO reference set when precision and recall are balanced. Precision is defined as the fraction of predicted ortholog pairs that are true, while recall is the fraction of true ortholog pairs that are predicted. Defined in terms of true positives (TP), false positives (FP), and false negatives (FN), precision (P) and recall (R) are:
$$P = {T_P \over T_P+F_P}$$ $$R= {T_P \over T_P+F_N}$$A brief description of each confidence score is as follows:
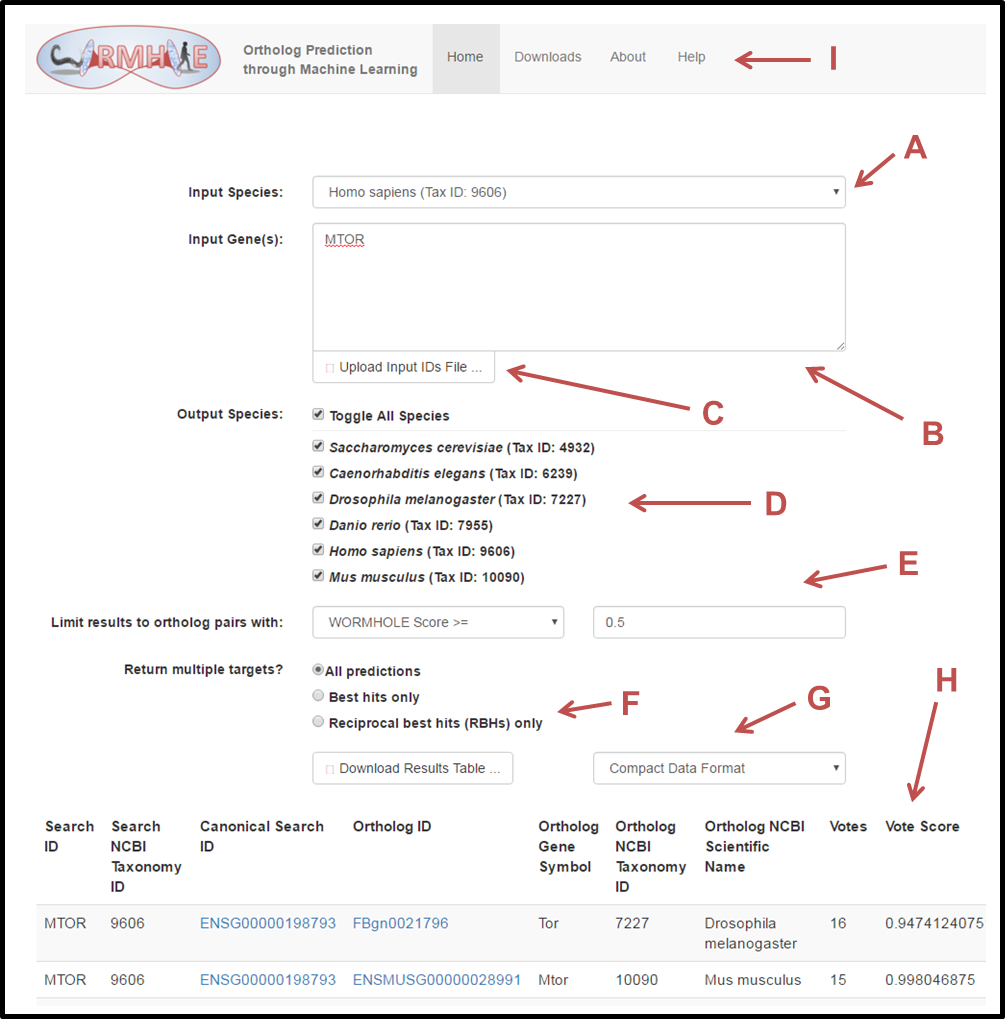
A. Input Species: The dropdown menu selects the query species that will be used for the ortholog prediction inquiry. Gene identifiers entered into the "Input Gene(s)" field (B) will search only the selected Input Species. WORMHOLE allows only one Input Species to be queried at a time.
B. Input Gene(s): Type or paste gene identifiers into this field to search for orthologs. Gene names or identifiers from the following sources are accepted by WORMHOLE:
WORMHOLE uses the Ensembl Gene ID from Ensembl version 77 as the primary identifier. If WORMHOLE does not produce a prediction using an identifier from a different source, we recommend that you retry the query using the Ensembl Gene ID for that gene. A complete list of gene aliases is available in the Download Data section of this website.
Each identifier should be separated by white space (i.e. space, tab, or new line). There is no "submit" button. The query will begin automatically as soon as you enter text into the input box. Ortholog predictions will appear at the bottom of the page (F). The number of IDs included in a single query is not limited; however, large queries will take time to load (e.g. a query consisting of 100 input genes will take approximately 2 minutes to complete). For very large queries, a complete list of genome-wide ortholog pairs is provided for immediate download in the Download Data section of this webpage.
C. Upload Input IDs File: As an alternative to using the text entry box, lists of gene identifiers can be uploaded in the form of a text file. Each identifier should be separated by white space (a space, tab, or new line).
D. Output Species: Check the boxes for which output species you wish to receive ortholog predictions. WORMHOLE allows multiple output species in the same query.
E. Score Filter: Ortholog queries can be filtered by one of several scores: Votes, Vote Score, or WORMHOLE Score. Selecting "Votes" will limit the output ortholog pairs to those predicted by at least the indicated number of input algorithms. Each of the other options places a minimum threshold on one of the three performance-scaled confidence scores calculated by the WORMHOLE multilayer machine learning models (see Overview above for details). By default, the filter is set to include all orthologs that achieve a WORMHOLE Score of at least 0.5. This value was selected because the WORMHOLE SVMs achieve optimal LDO prediction performance with balanced precision and recall at a threshold of 0.5. We recommend filtering queries with WORMHOLE Score >= 0.5 for casual queries.
F. Best Hits (BHs) and Reciprocal Best Hits (RBHs): By default, WORMHOLE will return all predicted genes pairs that include one of the queried genes (B) with Votes, Vote Score, or WORMHOLE Score above the selected threshold. A more stringent search can be performed by requesting only Best Hits or Reciprocal Best Hits. Selecting Best Hits will only return the target gene (or in a few cases, genes, when there is a tie) for each query gene that receivesthe highest WORMHOLE Score. Selecting Reciprocal Best Hits will further restrict the search by only returning a Best Hit target gene if the corresponding query gene is also the Best Hit for that target gene when the query is reversed. This is analagous to BLASP RBHs.
G. Download your results: This button is discontinued. If desired use copy-paste to transfer your web-based Results table to excel or similar.
H. Results table, Predicted Ortholog Pairs: As soon as text is entered into the input box or an input file is uploaded, the WORMHOLE query will begin automatically (this can take a few seconds for small queries, or several minutes for large queries). Ortholog predictions for the input genes will appear at the bottom of the page. The HTML output includes information about the input gene, output gene, output species, and the various WORMHOLE confidence scores. Clicking on the gene identifier will redirect you to the Ensembl page for that gene.
I. Download static data files: In addition to the web tool, several summary data files are available for download including genome-wide ortholog predictions for each pair of query and target species, a list of all aliases included in the WORMHOLE database for each gene, a list of all genes included in the WORMHOLE database, and the sets of reference ortholog pairs used to train and test the WORMHOLE SVMs.